February 2019
January
Nathaniel Guilford, graduate student
1 February 2019
Once again, my project has taken a turn in a new direction, but in my opinion it is down a very exciting route. Initially, we were going to need to source harbor seal tissues to obtain high quality DNA for sequencing. Using these reads, we would be able to identify polymorphic loci (SNPs) that could eventually be screened for in seal scat samples to obtain individual genotypes, thereby creating a non-invasive tracking method. While entirely feasible, the process of sourcing tissues from a wide range of the Eastern Pacific and gathering the proper authorization can take some time. This is why I was very happy to stumble upon a paper from January 2018 describing a new method that allowed us to look differently this ‘tissue-issue’. This method from Chiou and Bergey 2018, dubbed ‘FecalSeq’, allows for the direct next-gen sequencing of DNA straight from scat, which we already have a ton of! Previously, we had not planned on using scat DNA for sequencing due to its degraded nature and extremely high proportion of bacterial DNA. However, FecalSeq takes advantage of the difference in methylation patterns of bacterial and vertebrate DNA, and magnetic beads with bait proteins selectively bind only to vertebrate DNA in the fecal-extract. Washing away of the bacterial DNA leaves only the vertebrate DNA, of which the host DNA is a high enough proportion for sequencing (especially since we have the closely related Phoca largha genome to use as a reference).
While allowing us to employ an exciting new method, this also cuts down on future genotyping steps for us when validating our SNPs. Since we are using our scat samples to identify these markers, our sequence reads will also provide us with individual genotypes for our samples. If we had used tissues to identify these markers, we would have had to validate them on fecal extracted DNA as a first step, post SNP panel creation. With this method, we can begin to test scat DNA from both captive seals (where we know we will have re-sampled individuals as there are only a few seals in the same enclosure), as well as previously confirmed wild re-sampled individuals from Andrew Rothstein, a previous student of the lab using microsatellites. Any additional genotyping of wild scat samples would serve as more validation of the markers or an attempt to capture resampled individuals at a haul-out site, proving our tracking method viable.
Restricting our marker identification DNA pool to Salish Sea scat would not allow us to examine the population structure of many Eastern Pacific populations, however should we have time to ask additional questions, we could look at population differentiation within the range our scat samples do represent. Previous studies have found significant genetic differentiation within the inland Washington waters (Huber et al. 2012). Therefore, time permitting, we could compare the performance of SNPs in determining these population delineations compared to these previous studies which used either microsatellites or mtDNA.
Even if we do not get to population investigations, the new method for sequencing of fecal DNA and the subsequent testing of our new markers represents an exciting project that I look forward to developing. Now, to make these changes in my thesis proposal and begin putting it all together by the end of this quarter!
References:
- Chiou, K.L., and Bergey, C.M. (2018). Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces. Scientific Reports 8: 1975.
- Huber, H.R., Dickerson, B.R., Jeffries, S.J., and Lambourn, D.M. (2012). Genetic analysis of Washington State harbor seals (Phoca vitulina richardii) using microsatellites. Canadian Journal of Zoology 90, 1361–1369.
To record noise
Wyatt Heimbichner Goebel, undergraduate student
1 February 2019
I’m at the point in my project where I am starting to do trial runs so that I can refine my methodology. As such, most of my recent work in the lab has been focused on standardizing my methods such that the noise readings I take at each site are comparable as well as doing a lot of reading to see how other scientists have approached similar questions. Unfortunately, standardizing my methodology feels like a major challenge at this moment and I’m not quite sure how to proceed.
There are several things about trying to standardize noise measurements that make it difficult. The first of those is that the distance between the source of the noise and the observer measuring the noise may be different than the distance between the source and the seals. This is a problem because it could potentially cause the observer to record noise readings above or below the level that the seals are experiencing. This makes the vantage point of the observer a point of particular concern. I am trying to find vantage points that position the observer a similar distance from the seals at each site, but I haven’t quite settled on vantage points yet for Semiahmoo spit or Bellingham marina. I think that the issue of vantage point will be resolved with more trial runs. This problem also means that the observer must keep track of where the sound is coming from in order to keep the measurements as similar as possible. I’m not exactly sure how to handle this as it seems like the sources of noise are essentially unpredictable and could possibly change during the course of an observation. One potential course of action is to only compare noise measurements when the noise originates from a similar location relative to the observer and the seals, but this could potentially mean a lot of time investment in observations that yield unusable data and it is also unclear how I might determine if the various locations of noise sources are similar enough to warrant comparison.
Another challenge that I am having with respect to standardizing my methodology is the fact that the ambient noise level will change over the course of an observation. As such, it would be ideal to be able to continuously record the noise level throughout an observation, but this is not possible with the equipment that we currently have in the lab. I’m not sure how to handle this challenge yet either. One idea that I had was to have the observer take several readings throughout the course of the observation and then calculate the average noise level. However, the problem with this method is that any large increase in noise that occurs between noise readings will not be captured in the average, but could influence the number of seals hauled out. Clearly, I still have a lot of work to do with respect to finalizing my methods, but I hope to have these issues resolved within the next few weeks.
Refining my research question
Madison McKay, undergraduate student
1 February 2019
Because it will be difficult to find enough evidence of the parked behavior, and to be sure that is the behavior being used, I am working on refining my research question. I am now interested in investigating which factors best predict foraging success in harbor seals. I am planning to still look for interactions between recreational fishermen and the success of seals in the process. Some factors I am considering are: presence of fishermen (Y/N), tide, weather, number of seals present, and number of salmon.
The Whatcom Creek Hatchery records weekly return numbers of chum during their run in the Fall, and releases juveniles in April and May (Weekly Hatchery Escapement Report). I hypothesize that as the number of salmon increase, the foraging success of the seals will increase as well. This would mean we are seeing a larger amount of successful foraging in the creek during Fall and early Spring.
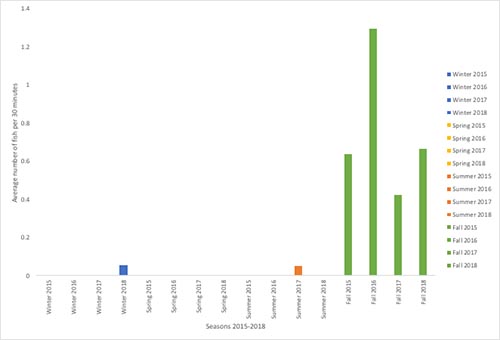
Figure 1. The average number of fish caught by seals per 30 minutes at Whatcom Creek, split into seasons as well as years.
In the figure above I calculated the average number of fish caught per 30 minutes by all observed seals. As we can see, Fall is the primary season for seals to successfully hunt. This makes sense given that the salmon run at Whatcom Creek occurs in the Fall. We can also notice that there were zero observed fish caught by the seals in Spring during all years. Because of this, I think it would be a good idea to focus on Fall and the variation of hunting success during this season and between years. I can also look into Winter 2018 and Summer 2017 in more detail. I did not include any information on the number of seals present in the figure above, which will be important in the future to determine the amount of fish being caught per seal.
From here, I am going to further investigate the importance of the factors I listed above. It is possible that one of these things, a combination, or all, are causing this variation in success from year to year. I am also excited to look at the hatchery data and see if the number of fish during the Fall seasons of 2015-2018 have any sort of correlation with the success rate of the seals.
January
Alisa Aist, undergraduate student
1 February 2019
Back in December I talked about if it would benefit the seals to remain in this area or if they should leave just because of the forecasted increase in human traffic in the Whatcom creek waterway. Now I want to pose the question of the impact of the seals in ecosystem. These seals don’t just stay in Bellingham Bay their whole lives they also go out into the greater Salish Sea and forage. A common food for harbor seals is salmon both as juveniles heading out to the ocean and as adults returning to their natal rivers to spawn (Thomas et al., 2017).
As many know the Southern Resident Killer Whales in the Puget Sound and San Juan Islands are endangered as their populations numbers are dropping and there are many stressors affecting them (Task Force, 2018). One of these stressors is the lack of their preferred food source, Chinook salmon. Now seals are not the only reason there maybe less Chinook in the Salish Sea but they are one reason.
This shows just how the harbor seal numbers can play a role in the Salish Sea ecosystem. All five pacific salmon species use the Nooksack river and there is a chum hatchery in Whatcom Creek. Salmon are currently being monitored in their behaviors at Whatcom Creek but not at the Nooksack, which has a much larger run. Seals also feed on squid and other fish species when there are not salmon around and can travel all over the Salish for food if needed.
As one of the top predators in Bellingham Bay if the harbor seals left there could be some changes in populations in the lower trophic levels. There could be an initial increase in fish numbers, but if the zooplankton are depleted then the fish will lose their food source and their numbers will go down as well. This increase decrease cycle does currently happen with seals in the ecosystem but the swings high and low are not as large as they would be without seals. There's also the possibility of some population collapses which would also have more cascading effects.
Overall I think that it is stabilizing for the seals to be a part of the Bellingham Bay ecosystem as long as the population stays at a reasonable level. Replacing the past haul out sites with new ones will not likely increase the population but rather maintain it at the same level as currently present.
References:
- Thomas AC, Nelson BW, Lance MM, Deagle BE, Trites AW (2017) Harbour seals target juvenile salmon of conservation concern. Canadian Journal of Fisheries and Aquatic Sciences 74:907-921.
- Task Force (2018) Southern Resident Orca: Report and Recommendations, Executive Order 18-02.
January
Jonathan Blubaugh, graduate student
3 February 2019
Winter quarter begins! I completed the outline for my proposal early this month. Outlining was harder than I thought. It was difficult organizing all the topics I want to include in a way that flows and makes sense. The methods section was the hardest part to outline because I am still learning how to use Ecopath. Writing methods for a technique that I don’t fully understand is a unique challenge. I am focused on learning more about how Ecopath works and the theory of ecosystem-based management so I can explain these concepts easily and succinctly in my proposal. The other sections of my proposal were easier to outline. The next step is translating the outline into a real proposal which is even more daunting than writing the outline itself.
Teaching has also come back with the start of winter quarter. So far it seems to be going easier than last quarter, with no major hiccups so far. Georgianne observed me last week and I think it went well. I am always more nervous when someone is observing me teach. I am optimistic that I will enjoy teaching a bit more this quarter and hopefully I can dedicate more time to research because I won’t be as stressed about teaching.