Cougar population structure and spatial genetic
variation
in
Washington and southern British Columbia*
Matthew Warren and
David Wallin
Department of Environmental Sciences
Huxley College of the Environment
Western Washington University
Bellingham, WA 98229
Richard Beausoleil
Washington Department of Fish and Wildlife
3515 State Highway 97A
Wenatchee, WA 98801
*This webpage is based on a poster that was presented at the Oregon and Washington Chapters of the Wildlife Society 2013 Annual Meeting; 2013 February 13-15; Stevenson, WA. (Winner best graduate student poster)

Background
In Washington,
cougar mortality may not affect regional population density if it is offset by
increased immigration of subadult males from
surrounding areas (Robinson et al. 2008). Effective management of cougars
therefore requires a better understanding of population structure across the
state, specifically whether the stateís cougars comprise a single, panmictic population, or function as a metapopulation.
The primary aim of
this study was to describe patterns of variation in microsatellite allele
frequencies across the state, in order to reveal the underlying population
structure in the region. We also looked for evidence of male-biased dispersal,
to evaluate the contribution of sex-biased dispersal to population structure.
Methods
The Washington
Department of Fish and Wildlife (WDFW) obtained 667 muscle and tissue samples
from cougars across Washington and southern British Columbia between 2003 and 2010.
Polymerase chain reaction was used to amplify 17 previously characterized
microsatellite markers.
We used the Bayesian
clustering program Geneland to estimate the number of
genetic clusters within the study area and assign individuals to those clusters
(Guillot et al. 2005). Spatial PCA was also used to
visualize genetic patterns (Jombart et al. 2008).
We looked for
evidence of male-biased dispersal in genotype frequencies using an assignment
index (AIc) test in Fstat,
which calculates how likely a given genotype is to occur within that population
(Goudet et al. 2002).
We tested for an
effect of geographic distance on genetic distance using a simple Mantel test.
We used principal components analysis (PCA) of allele frequencies to calculate
genetic distances between individuals (Shirk et al. 2010).
Results
Support was highest
for four populations in the study area in the Geneland
simulations (Figure 1). The first global sPCA
axis differentiated the northeastern cluster from the Cascades cluster (Figure
2a), while the second global axis separated the Olympic and Blue Mountain
clusters from neighboring clusters (Figure 2b). Population differentiation (FST)
increased with distance between clusters (Table 1). Genetic distance was
positively correlated with geographic distance, however the relationship was
fairly weak (r = 0.33, P = 0.001).
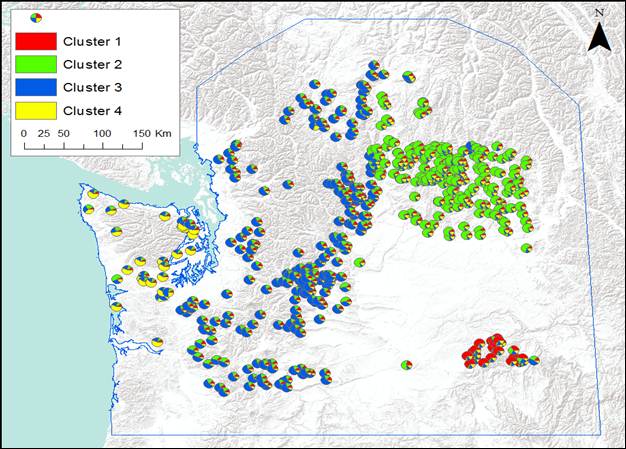
Figure 1. Posterior probability of membership in Geneland clusters for individual cougars; samples collected
between 2003-2010.
Table 1. Estimates of genetic
differentiation (FST) between cougar population clusters.
|
Blue Mtns |
Northeast |
Cascades |
|
|
Northeast |
0.094 |
-- |
-- |
|
Cascades |
0.151 |
0.036 |
-- |
|
Olympic |
0.310 |
0.205 |
0.145 |
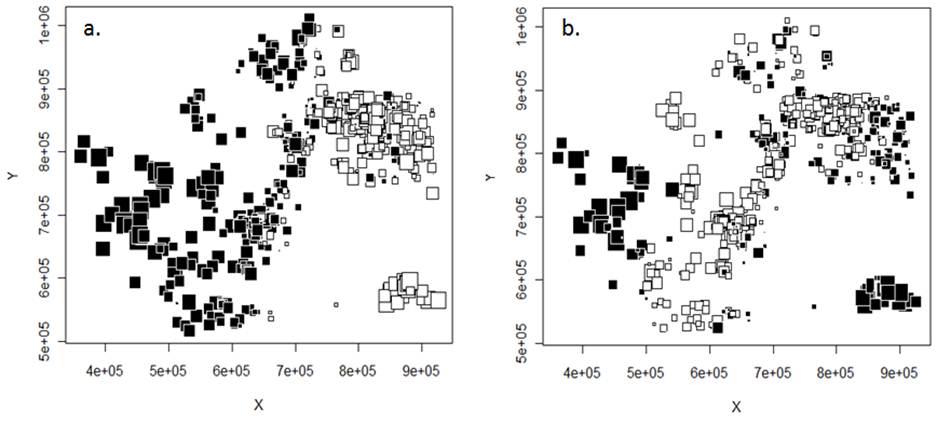
Figure 2. First (a) and second (b) global sPCA axes.† Genetic similarity is represented by color and size of squares, where squares of different color are strongly differentiated from each other, while squares of similar color but different size are weakly differentiated.† Geographic coordinates in UTMís are shown on the X and Y axes.
The mean assignment
index (AIc) value was significantly lower for males
than for females, as would be expected with male-biased dispersal (P = 0.029;
Table 2). Also in keeping with male-biased dispersal, male cougars exhibited
significantly higher FIS values than female cougars (P = 0.003;
Table 2). The variance in AIc values and FST
values were not significantly different between the sexes (Table 2).
Table 2. Permutation test results for sex-biased dispersal in cougars. Results in bold are significant at α=0.05.
|
n |
Mean AIc |
Variance AIc |
FIS |
FST |
|
|
Female |
301 |
0.388 |
25.02 |
0.0297 |
0.0629 |
|
Male |
308 |
-0.379 |
30.04 |
0.0761 |
0.0831 |
|
P-value |
0.029 |
0.121 |
0.003 |
0.188 |
Conclusions
My results suggest that cougar populations in Washington and southern British Columbia are structured as a metapopulation and not a single, panmictic population. The results of Geneland clustering largely agreed with those of spatial PCA, showing four clusters in the study area, with an area of mixed genotypes in north-central Washington along the Okanogan Valley.
Given an average dispersal distance of 190-250 km for male cougars in Washington, the degree of differentiation between clusters is surprising. The weak relationship between genetic distance and geographic distance suggests that isolation by distance is not solely responsible for the observed pattern of genetic differentiation. Further research will explore the relationship between landscape characteristics and gene flow in northwestern cougars.
Results for all four dispersal tests were in line with male-biased dispersal, however the variance of AIc and FST tests were non-significant. Goudet et al. (2002) found that the variance of AIc test performs best when dispersal rates are <10%; the propensity for male cougars to disperse from natal areas may have resulted in low power for this test.
Acknowledgements
We would like to thank the Washington Department of Fish and Wildlife, who collected and analyzed all cougar DNA samples.† We would also like to thank Spencer Houck and Heidi Rodenhizer for GIS assistance, and Dietmar Schwarz for help with data analysis.† Funding was provided by Washington Department of Fish and Wildlife, Western Washington University, Seattle City Lightís Wildlife Research Grants Program, the Washington Chapter of the Wildlife Society, and North Cascades Audubon Society.
References
Goudet, J., N. Perrin, and P. Waser. 2002. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology 11: 1103-1114.
Guillot, G., A. Estoup, F. Mortier, and J.F. Cosson. 2005. A spatial statistical model for landscape genetics. Genetics 170: 1261-1280.
Jombart, T., S. Devillard, A.B. Dufour, and D. Pontier. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101: 92-103.
Robinson, H.S., R.B. Wielgus, H.S. Cooley, and S.W. Cooley. 2008. Sink populations in carnivore management: cougar demography and immigraton in a hunted population. Ecological Applications 18: 1028-1037.
Shirk, A.J., D.O. Wallin, S.A. Cushman, C.G. Rice, and K.I. Warheit. 2010. Inferring landscape effects on gene flow: a new model selection framework. Molecular Ecology 19: 3603-3619.