NOVEMBER 2019
Grace's blog
Grace Freeman, graduate student
1 November 2019
What a whirlwind! I can’t believe it’s already time to write another monthly blog. Before starting at Western, I was told that the quarter system moved quickly, and that I needed to stay on top of things to keep up, and now I certainly believe it!
TAing has definitely been the thing taking up the majority of my mental bandwidth these days. I’m teaching lab for human physiology, a class I’ve never taken, which means I have to learn a lot about the human body before lecturing on it each week. I look forward to next quarter when I will be teaching the same course; I’m hoping it’ll feel like a breeze since I will already know what to expect!
Unfortunately, learning to balance classes, TAing, and Washington life in general has meant that my thesis proposal has fallen to the side a bit. I must admit that Alejandro was right, and I am glad to have put in the work over the summer, so I don’t have to do it all now. That said, I’m still eager to dive back in. I haven’t worked on the proposal itself this month but submitting an NSF GRFP application has certainly set me up well and helped to clarify my thinking and questions. I’m excited to take what I’ve learned from that process and apply it back to my thesis.
On another note, I need to have a committee meeting before the end of the quarter, and to do that, I need a committee! As part of one of my classes, Bio 521, we have to meet with and interview several faculty members from Western. The goal is that I will be able to ask a few of these people to formally help me on the project and serve as committee members. This process has been so helpful in opening the channels of communication and introducing me to the expertise these people may be able to provide. I’m hoping that in next month’s blog I will be able to report a successful search. Until then, if you need me, I will be busy writing and meeting with bio faculty!
Blog Post 2
Bobbie Buzzell, graduate student
1 November 2019
While I’ve settled into the routine and pace of graduate school, scat cleaning and processing continues. Earlier this month, I made a trip out to Neah Bay to pick up the remaining scat samples collected in the month of September. The final counts are in-539 scats!
Although I have cleaned more than 200 scats, I still have much more to go which will be my primary focus this quarter and during winter break. I can clean approximately 15 scats in about 3 hours, and this is done through washing the scat through 1 (or 2 if needed) nested mesh sieves. Forceps are used to recover all of the fish bones and invertebrate remains, and then placed in vials with alcohol. After a couple of weeks, the vials are placed in a drying oven for 3 days to evaporate all the liquid. This provides a much more pleasant experience when working with the samples later during the identification stage.
In the final week of October, I have readied a few samples to be sent to William Walker, a food habits specialist at the National Marine Mammal Laboratory in Seattle. He will be completing a preliminary analysis of the fish found in these samples. After this assessment, we will have a good idea of how long the prey identification will take with each sample. Currently he estimates a half hour per sample to identify fish prey. With 539 samples and a 40 hour work week, this works out to approximately 7 weeks of fish prey identification. Time is of the essence for this project, so it’s important I meet those benchmarks over the next few months.
You’ll find me in the basement, in the scat cleaning room.
October Blog
Jonathan Blubaugh, graduate student
1 November 2019
Results! After many months of working on my Ecopath model, I finally have some preliminary results to share. I used the Monte Carlo sampling routine in Ecosim to generate 10 balanced models with male and female harbor seals in their own functional groups. Then I used the RPath package in R, to reapportion the seal biomass for each sex ratio, from 10% females to 90% females in 5% increments. This created 170 models, 10 for each of the 17 sex ratios. Then each model had the Mixed Trophic Impact (MTI) calculated, which is a measure of the impact a predator group has on a prey group including competition and trophic cascade effects. I extracted the MTI for the male and female harbor seals and graphed their impacts on the salmon groups below.
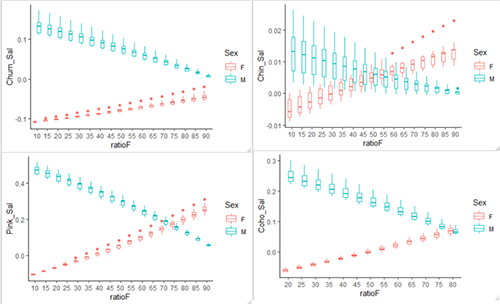
Mixed trophic impact on four species of salmon.
These preliminary results show the differences in impact that male and female harbor seals could have on their prey items. This is more than just differences in consumption since male harbor seals tended to have a greater positive impact on salmon compared to females, even though male seals eat more salmon then females. This holds true even when there is a large proportion of females in the model.
I’m really excited to explore these data more since there appear to be some really interesting interactions and the results will be different for each impacted group. Next month will have more in-depth analysis of these interactions!
October Blog
Nathan Guilford, graduate student
1 November 2019
This month brought the good news that all fifteen of my enriched DNA samples have made it through library preparation and are ready to be sequenced! We have since discussed the option to increase our sequencing coverage from ~two to ~four million reads per sample by investing in a half-lane on an Illumina NextSeq, but decided to stick with the original quarter-lane in order to save funds for a second sequencing run should that be necessary. Due to the enrichment process performed on these samples removing (hopefully) most of the bacterial DNA, we believe that this sequencing depth should be adequate for distinguishing individuals. Additionally, we may achieve slightly deeper sequencing than expected, due to the removal of two of the fifteen samples prior to sequencing. Within the fifteen samples are five replicates from the captive seal, while only three are needed. Now, the remaining thirteen samples will be sequenced, and I will soon have data to explore and analyze!
I was also recently able to present my work to the board of the College of Science and Engineering this month. This summer, Dr. Acevedo-Gutierrez received the Arlan Norman Award for Excellence in Student Mentoring, an award which granted me (as his graduate student) generous support. As a recipient of this funding, I was more than happy to show what I have been working on to the college and personally thank both the CSE and Dr. Norman.
October 2019
Delaney Adams, undergraduate student
1 November 2019
Happy Halloween!! It’s been such a nice surprise to have a mostly sunny last couple of weeks, but it is especially exciting to me to have a sunny Halloween day. November has snuck up on me rather quickly, and I feel as if October has been just a blur. The time has finally come that I have made a decision on what I am going to be focusing my project on throughout the year. Much conversation, contemplation, and swinging back and forth on what direction I wanted to take has finally led me to a decision. Regardless, I am going to be focusing on harbor seal hunting success throughout the remainder of the year, continuing Madi’s project on the relationship between seals and the sports fishermen that occupy Whatcom creek throughout most of the fall months. Last year, she focused on that and determined that the number of seals was the most predictive indicator of the hunting success in the creek, rather than fishermen, and I am going to continue that with more recent data and will try to find out more about what is going on there (are the seals hunting cooperatively or is the number of seals in the creek just an indication of the abundance of the fish as well?). I am starting by making an outline with just general ideas for the main sections of this project, and I will be working on that throughout most of the month, as well as writing a proposal for my Honor’s senior project, which will be encompassed in this project for Whatcom creek lab.
Outside of the lab, school is keeping me extraordinarily busy! My classes are proving to be quite challenging, work-intensive, yet also extremely rewarding. On top of that, I am involved in sports, clubs, and have a new job on campus that gets me out and about, providing me with ample opportunities to explore campus life. I am enjoying it all, but am also definitely looking forward to everything calming down here in the near future.
Update on my Independent Research Project
Helen Krueger, undergraduate student
1 November 2019
Wheeww! It has been a month of toying with the idea of researching microbial community growing on haul out logs in the log pond and Whatcom waterway. I originally wanted to sequence the V4 16S region of microbial rRNA and compare OTUs between differing sites. But upon talking to several more informed professors/professionals, I realized that the cost and time investment was much more than what I could handle by the spring deadline.
Still intrigued by the idea of seal haul-outs harboring more microbes than a control log with no seals on them, I decided to adjust the scope of my proposed study. By talking with Dr. Craig Moyer, who is well versed in DNA sequences, we came to the conclusion that quantifying the DNA would be better. Because Dr. Moyer has never worked with a wood substrate before, and I had many questions about isolating and extracting the microbial DNA, I went to Google Scholar to look into how to go about extraction.
The best experimental procedure, that I found, was from an article on wood colonizing bacteria (Proença 2017). 5-gram shavings of wood were ‘shook’ in NaCl at 25 degrees Celcius for 2 hours (Proença 2017). The samples were then frozen or directly isolated. This was to ensure that the microbes were suspended in the solution instead of being integrated with the wood sample (Proença 2017). For isolation, the sample should be serially diluted and then plated onto agar, grown on agar for 5 days in a dark environment (Proença 2017). Using microscopy to count individual colonies will act as one quantifying test. Additionally, I will use the Qubit 2.0 and Qubit Fluorometer to quantify serially diluted samples (McPherson, 2018).
There are still many many kinks to work out before I am ready to get begin sampling and quantifying, but I think that I am finally on the right track.
References:
- McPherson, M. R., Wang, P., Marsh, E. L., Mitchell, R. B., & Schachtman, D. P. 2018. Isolation and analysis of microbial communities in soil, rhizosphere, and roots in perennial grass experiments. Journal of Visualized Experiments 137: 57932.
- Proença, D. N., Francisco, R., Kublik, S., Schöler, A., Vestergaard, G., Schloter, M., & Morais, P. V. 2017. The microbiome of endophytic, wood colonizing bacteria from pine trees as affected by pine wilt disease. Scientific Reports 7: 4205.